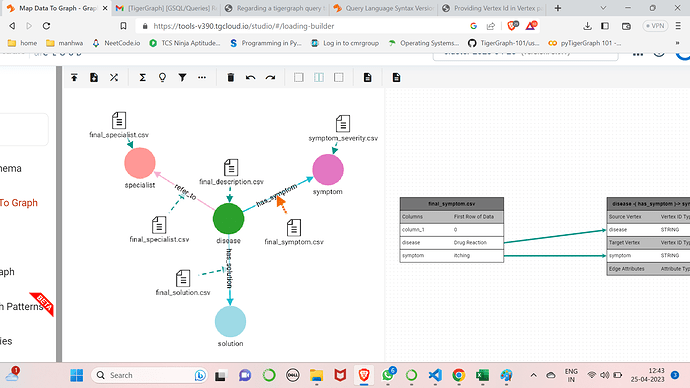
I have 2 vertexes one called disease and other called symptoms and an undirected edge has_symptom connecting the two.Now how do i write a query such that the user gives a set of symptoms as strings and the query can provide a disease name which matches with the given symptoms.
Points to note -
for each disease the symptoms are stored in the vertex id of the symptom vertex.
Please do help me with the query
probably something like this , I used SYNTAX V2
CREATE QUERY findDisease(SET<STRING> symptomsArg) FOR GRAPH yourGraphName SYNTAX V2 {
SYM1 = SELECT s FROM symptom:s where s.symptomName IN symptomsArg;
DIS1 = SELECT d FROM SYM1 -(has_symptom)- disease:d;
PRINT DIS1;
}
I am getting an empty list.Following are the points i noted for my graph
Expect for one symptom the rest of them give an empty list
This is the schema i am using along with the has_symptom edge mapping.
The symptom vertex has another file mapped with attributes name(Primary ID) and severity and i dont make use of the severity anywhere
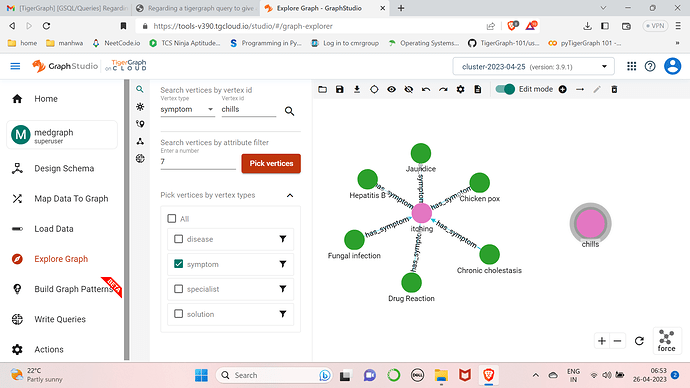
@Bhuvanesh Can you go to the “Load Data” tab to confirm that the data was loaded? Also if you go to “Explore Graph” you can manually search for data to confirm it was loaded correctly. I would start there. After that, it might be due to the query being run.
Continuing the discussion from Regarding a tigergraph query to give a matching vertex:
@Jon_Herke I checked the Load data lab and there all the files are loaded without errors
I tried using a directed edge from disease to symptoms now.When i double click on itching it actually shows the directed edges from the particular disease but the same isnt happening for the rest of the symptoms.
@Bhuvanesh Hey mate you can do it like this to find a disease that matches almost all of the given symptoms, you can create a query that searches for diseases connected to each symptom. For each symptom, gather the connected diseases. Then, count how many times each disease appears. Finally, identify the disease that appears the most and return its name as the result. This way, you’ll get the disease that is connected to the most input symptoms.
i hope this will help you